Janies applies sequencing, supercomputing to correct erroneous classification
Columbus, Ohio (April 27, 2011) – A team of scientists has combined embryological observations, genetic sequencing, and supercomputing to determine that a group of small disk-shaped animals that were once thought to represent a new class of animals are actually starfish that have lost the large star-shaped, adult body from their life cycle.
In a paper for the journal Systematic Biology (sysbio.oxfordjournals.org), Daniel Janies, Ph.D., a computational biologist in the department of Biomedical Informatics at The Ohio State University (OSU), leveraged computer systems at the Ohio Supercomputer Center (OSC) to help support his contention that class-level status of Xyloplax does not reflect their evolutionary history.
“Although Xyloplax does not represent a new class, it an even more interesting animal now because it represents a rare example of how natural selection can shape the whole the life cycle,” he explained. “By omitting the large adult stage, Xylopax found how to make a living in the nooks and crannies of sunken timbers on the deep-sea floor.”
Janies collaborated on the paper with co-authors Janet R. Voight, Ph.D., in the department of Zoology at the Field Museum of Natural History in Chicago, and Marymegan Daly, Ph.D., in the department of Evolution, Ecology and Organismal Biology at Ohio State.
Janies and his colleagues are examining echinoderms – starfish, sea urchins and their close relatives – as part of a study for the United States’ National Science Foundation’s Assembling the Tree of Life project. Several OSU scientists, including Janies and Daly, have won these grants, typically valued at $3 million over five years, to better understand the interrelationships of all forms of life (http://echinotol.org).
As tree-of-life analyses are computationally difficult, the current project is enabled by the computational muscle of OSC’s flagship system, the 9,500-node IBM 1350 Opteron “Glenn Cluster.”
 |
| Daniel Janies, PhD. |
“Last year, OSC deployed a $4 million expansion to the Glenn Cluster with a specific focus on supporting researchers, like Drs. Janies and Daly, in Ohio’s growing biosciences sector,” said Ashok Krishnamurthy, interim co-executive director of OSC. “The center provides a world-class computing environment for amazing bioscience research projects at places like OSU’s Department of Biomedical Informatics, the Research Institute at Nationwide Children’s Hospital and the Cleveland Clinic.”
The researchers examined both genetic and anatomical data to support their hypothesis, comparing Xyloplax with a total of 86 species representing major lineages of the five living classes of echinoderms (sea cucumbers, sea urchins, starfish, brittle stars, and sea lilies). For accurate comparisons, they applied several analytical methods to search for the shared mutations and changes in anatomy among the lineages.
The results describe the relationships in a phylogenetic tree representing the best-supported hypothesis on how Xyloplax evolved from other echinoderms. Phylogenetics is the study of the evolutionary relationships and changes among various biological species as they evolve from a common ancestor.
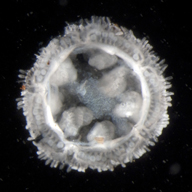
Prior to this study, molecular investigations of Xyloplax were limited, because specimens in two early collections in the 1980s were fixed in a solution which degrades DNA. Voight recently collected specimens of Xyloplax in the northeastern Pacific Ocean and carefully preserved them in ethanol. From these specimens, Daly was able to sequence several genes for Xyloplax. The newer specimens of Xyloplax also included several brooding females containing embryos, providing Janies with an unprecedented view of the creature’s early development.
Janies and colleagues theorize that Xyloplax differs from other starfish species because it is progenetic – that is, it has a rare, truncated life cycle that leaves the mature organism with features retained from its juvenile stages. For example, the arms of a starfish typically grow axially, like spokes of a wheel, as they develop from juveniles to adults, whereas Xyloplax grows along its circumference, like the wheel itself, and never develops arms.
“Irrespective of method or data sampling scheme, our results show that Xyloplax evolved from within starfish,” Janies concluded. “Xyloplax is just a little starfish that has a strange body plan and habitat, so strange that many could not recognize it as a starfish until we unlocked its genome and development.”
The Systematic Biology article is available online at:
http://sysbio.oxfordjournals.org/cgi/content/full/syr044? ijkey=JFlq6Bz3WkOSNqH&keytype=ref
The Ohio Supercomputer Center (OSC) addresses the rising computational demands of academic and industrial research communities by providing a robust shared infrastructure and proven expertise in advanced modeling, simulation and analysis. OSC empowers scientists with the vital resources essential to make extraordinary discoveries and innovations, partners with businesses and industry to leverage computational science as a competitive force in the global knowledge economy, and leads efforts to equip the workforce with the key technology skills required to secure 21st century jobs. For more, visit www.osc.edu.