Ascend
OSC's original Ascend cluster was installed in fall 2022 and is a Dell-built, AMD EPYC™ CPUs with NVIDIA A100 80GB GPUs cluster. In 2025, OSC expanded HPC resources on its Ascend cluster, which features additional 298 Dell R7525 server nodes with AMD EPYC 7H12 CPUs and NVIDIA A100 40GB GPUs.
Hardware
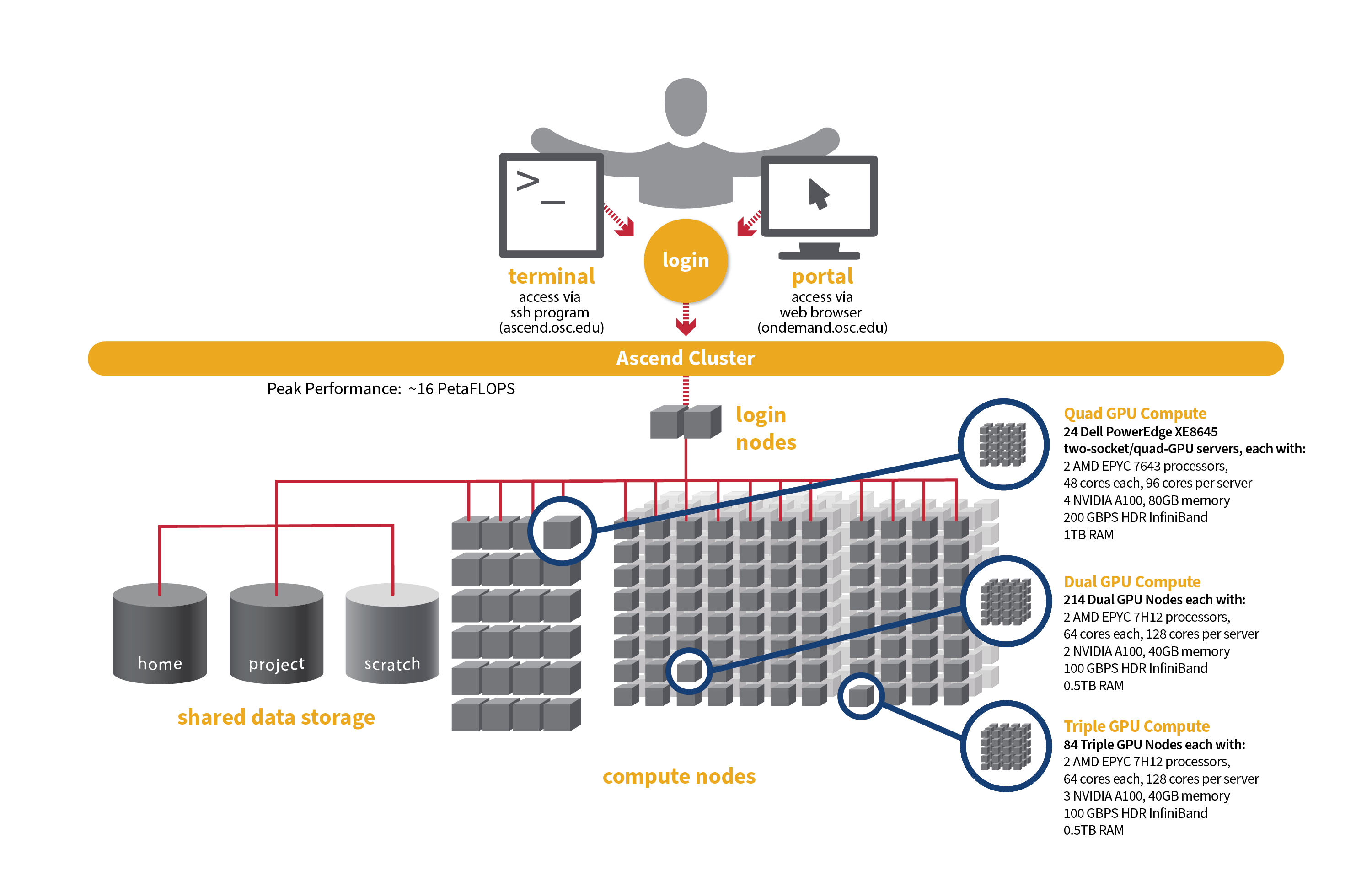
- Quad GPU Compute: 24 Dell PowerEdge XE8645 two-socket/quad-GPU servers, each with:
- 2 AMD EPYC 7643 (Milan) processors (2.3 GHz, each with 44 usable cores)
- 4 NVIDIA A100 GPUs with 80GB memory each, connected by NVIDIA NVLink
- 921GB usable memory
- 12.8TB NVMe internal storage
- HDR200 Infiniband (200 Gbps)
- Dual GPU Compute: 214 Dell PowerEdge R7545 two-socket/dual GPU servers (190 available to Slurm and 24 reserved for Kubernetes), each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 2 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
- Triple GPU Compute: 84 Dell PowerEdge R7545 two-socket/dual GPU servers, each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 3 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W (24 nodes with three GPUs are currently available. The remaining 60 nodes are still undergoing testing on their third GPU, so that GPU is not yet available for user jobs).
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
- Theoretical system peak performance
- ~16 PetaFLOPS
- 40,448 total cores and 776 GPUs (some cores and GPUs are reserved for Kubernetes)
- 2 login nodes
- IP address: 192.148.247.[180-181]
How to Connect
-
SSH Method
To login to Ascend at OSC, ssh to the following hostname:
ascend.osc.edu
You can either use an ssh client application or execute ssh on the command line in a terminal window as follows:
ssh <username>@ascend.osc.edu
You may see a warning message including SSH key fingerprint. Verify that the fingerprint in the message matches one of the SSH key fingerprints listed here, then type yes.
From there, you are connected to the Ascend login node and have access to the compilers and other software development tools. You can run programs interactively or through batch requests. We use control groups on login nodes to keep the login nodes stable. Please use batch jobs for any compute-intensive or memory-intensive work. See the following sections for details.
-
OnDemand Method
You can also login to Ascend at OSC with our OnDemand tool. The first step is to log into OnDemand. Then once logged in you can access Ascend by clicking on "Clusters", and then selecting ">_Ascend Shell Access".
Instructions on how to connect to OnDemand can be found at the OnDemand documentation page.
File Systems
Ascend accesses the same OSC mass storage environment as our other clusters. Therefore, users have the same home directory as on the old clusters. Full details of the storage environment are available in our storage environment guide.
Software Environment
The Ascend cluster is now running on Red Hat Enterprise Linux (RHEL) 9, introducing several software-related changes compared to the RHEL 7/8 environment. These updates provide access to modern tools and libraries but may also require adjustments to your workflows. You can stay updated on the software packages available on Ascend by viewing Available software list on Next Gen Ascend.
Key change
A key change is that you are now required to specify the module version when loading any modules. For example, instead of using module load intel, you must use module load intel/2021.10.0. Failure to specify the version will result in an error message.
Below is an example message when loading gcc without specifying the version:
$ module load gcc Lmod has detected the following error: These module(s) or extension(s) exist but cannot be loaded as requested: "gcc". You encountered this error for one of the following reasons: 1. Missing version specification: On Ascend, you must specify an available version. 2. Missing required modules: Ensure you have loaded the appropriate compiler and MPI modules. Try: "module spider gcc" to view available versions or required modules. If you need further assistance, please contact oschelp@osc.edu with the subject line "lmod error: gcc"
Batch Specifics
Refer to this Slurm migration page to understand how to use Slurm on the Ascend cluster.
Using OSC Resources
For more information about how to use OSC resources, please see our guide on batch processing at OSC. For specific information about modules and file storage, please see the Batch Execution Environment page.
Ascend Programming Environment
Compilers
C, C++ and Fortran are supported on the Ascend cluster. Intel, oneAPI, GNU Compiler Collectio (GCC) and AOCC are available. The Intel development tool chain is loaded by default. Compiler commands and recommended options for serial programs are listed in the table below. See also our compilation guide.
The Rome/Milan processors from AMD that make up Ascend support the Advanced Vector Extensions (AVX2) instruction set, but you must set the correct compiler flags to take advantage of it. AVX2 has the potential to speed up your code by a factor of 4 or more, depending on the compiler and options you would otherwise use. However, bear in mind that clock speeds decrease as the level of the instruction set increases. So, if your code does not benefit from vectorization it may be beneficial to use a lower instruction set.
In our experience, the Intel compiler usually does the best job of optimizing numerical codes and we recommend that you give it a try if you’ve been using another compiler.
With the Intel/oneAPI compilers, use -xHost and -O2 or higher. With GCC, use -march=native and -O3.
This advice assumes that you are building and running your code on Ascend. The executables will not be portable. Of course, any highly optimized builds, such as those employing the options above, should be thoroughly validated for correctness.
| LANGUAGE | INTEL | GCC | ONEAPI |
|---|---|---|---|
| C | icc -O2 -xHost hello.c | gcc -O3 -march=native hello.c | icx -O2 -xHost hello.c |
| Fortran | ifort -O2 -xHost hello.F | gfortran -O3 -march=native hello.F | ifx -O2 -xHost hello.F |
| C++ | icpc -O2 -xHost hello.cpp | g++ -O3 -march=native hello.cpp | icpx -O2 -xHost hello.cpp |
Parallel Programming
MPI
OSC systems use the MVAPICH implementation of the Message Passing Interface (MPI), optimized for the high-speed Infiniband interconnect. MPI is a standard library for performing parallel processing using a distributed-memory model. For more information on building your MPI codes, please visit the MPI Library documentation.
MPI programs are started with the srun command. For example,
#!/bin/bash
#SBATCH --nodes=2
#SBATCH --ntasks-per-node=48
srun [ options ] mpi_prog
The srun command will normally spawn one MPI process per task requested in a Slurm batch job. Use the --ntasks-per-node=n option to change that behavior. For example,
#!/bin/bash #SBATCH --nodes=2 #SBATCh --exclusive # Use the maximum number of CPUs of two nodes srun ./mpi_prog # Run 8 processes per node srun -n 16 --ntasks-per-node=8 ./mpi_prog
The table below shows some commonly used options. Use srun -help for more information.
| OPTION | COMMENT |
|---|---|
--ntasks-per-node=n |
number of tasks to invoke on each node |
-help |
Get a list of available options |
srun in any circumstances.OpenMP
The Intel, and GNU compilers understand the OpenMP set of directives, which support multithreaded programming. For more information on building OpenMP codes on OSC systems, please visit the OpenMP documentation.
An OpenMP program by default will use a number of threads equal to the number of CPUs requested in a Slurm batch job. To use a different number of threads, set the environment variable OMP_NUM_THREADS. For example,
#!/bin/bash #SBATCH --ntasks-per-node=8 # Run 8 threads ./omp_prog # Run 4 threads export OMP_NUM_THREADS=4 ./omp_prog
Interactive job only
Please use -c, --cpus-per-task=X to request an interactive job. Both result in an interactive job with X CPUs available but only the former option automatically assigns the correct number of threads to the OpenMP program.
Hybrid (MPI + OpenMP)
An example of running a job for hybrid code:
#!/bin/bash #SBATCH --nodes=2 #SBATCH --ntasks-per-node=80 # Run 4 MPI processes on each node and 40 OpenMP threads spawned from a MPI process export OMP_NUM_THREADS=40 srun -n 8 -c 40 --ntasks-per-node=4 ./hybrid_prog
Tuning Parallel Program Performance: Process/Thread Placement
To get the maximum performance, it is important to make sure that processes/threads are located as close as possible to their data, and as close as possible to each other if they need to work on the same piece of data, with given the arrangement of node, sockets, and cores, with different access to RAM and caches.
While cache and memory contention between threads/processes are an issue, it is best to use scatter distribution for code.
Processes and threads are placed differently depending on the computing resources you requste and the compiler and MPI implementation used to compile your code. For the former, see the above examples to learn how to run a job on exclusive nodes. For the latter, this section summarizes the default behavior and how to modify placement.
OpenMP only
For all three compilers (Intel, GCC and oneAPI), purely threaded codes do not bind to particular CPU cores by default. In other words, it is possible that multiple threads are bound to the same CPU core.
The following table describes how to modify the default placements for pure threaded code:
| DISTRIBUTION | Compact | Scatter/Cyclic |
|---|---|---|
| DESCRIPTION | Place threads close to each other as possible in successive order | Distribute threads as evenly as possible across sockets |
| INTEL/ONEAPI | KMP_AFFINITY=compact | KMP_AFFINITY=scatter |
| GCC | OMP_PLACES=sockets[1] | OMP_PROC_BIND=true OMP_PLACES=cores |
- Threads in the same socket might be bound to the same CPU core.
MPI Only
For MPI-only codes, MVAPICH first binds as many processes as possible on one socket, then allocates the remaining processes on the second socket so that consecutive tasks are near each other. Intel MPI and OpenMPI alternately bind processes on socket 1, socket 2, socket 1, socket 2 etc, as cyclic distribution.
For process distribution across nodes, all MPIs first bind as many processes as possible on one node, then allocates the remaining processes on the second node.
The following table describe how to modify the default placements on single node for MPI-only code with the command srun:
| DISTRIBUTION (single node) |
Compact | Scatter/Cyclic |
|---|---|---|
| DESCRIPTION | Place processs close to each other as possible in successive order | Distribute process as evenly as possible across sockets |
| MVAPICH[1] | Default | MVP_CPU_BINDING_POLICY=scatter |
| INTEL MPI | SLURM_DISTRIBUTION=block:block srun -B "2:*:1" ./mpi_prog |
SLURM_DISTRIBUTION=block:cyclic srun -B "2:*:1" ./mpi_prog |
| OPENMPI | SLURM_DISTRIBUTION=block:block srun -B "2:*:1" ./mpi_prog |
SLURM_DISTRIBUTION=block:cyclic srun -B "2:*:1" ./mpi_prog |
MVP_CPU_BINDING_POLICYwill not work ifMVP_ENABLE_AFFINITY=0is set.
To distribute processes evenly across nodes, please set SLURM_DISTRIBUTION=cyclic.
Hybrid (MPI + OpenMP)
For hybrid codes, each MPI process is allocated a number of cores defined by OMP_NUM_THREADS, and the threads of each process are bound to those cores. All MPI processes, along with the threads bound to them, behave similarly to what was described in the previous sections.
The following table describe how to modify the default placements on a single node for Hybrid code with the command srun:
| DISTRIBUTION (single node) |
Compact | Scatter/Cyclic |
|---|---|---|
| DESCRIPTION | Place processs as closely as possible on sockets | Distribute process as evenly as possible across sockets |
| MVAPICH[1] | Default | MVP_HYBRID_BINDING_POLICY=scatter |
| INTEL MPI[2] | SLURM_DISTRIBUTION=block:block | SLURM_DISTRIBUTION=block:cyclic |
| OPENMPI[2] | SLURM_DISTRIBUTION=block:block | SLURM_DISTRIBUTION=block:cyclic |
Summary
The above tables list the most commonly used settings for process/thread placement. Some compilers and Intel libraries may have additional options for process and thread placement beyond those mentioned on this page. For more information on a specific compiler/library, check the more detailed documentation for that library.
GPU Programming
244 NVIDIA A100 GPUs are available on Ascend. Please visit our GPU documentation.
Reference
Ascend Software Environment
The Next Gen Ascend (hereafter referred to as “Ascend”) cluster is now running on Red Hat Enterprise Linux (RHEL) 9, introducing several software-related changes compared to the RHEL 7/8 environment used on the Pitzer and original Ascend cluster. These updates provide access to modern tools and libraries but may also require adjustments to your workflows. Key software changes and available software are outlined in the following sections.
Updated Compilers and Toolchains
The system GCC (GNU Compiler Collection) is now at version 11. Additionally, newer versions of GCC and other compiler suites, including the Intel Compiler Classic and Intel oneAPI, are available and can be accessed through the modules system. These new compiler versions may impact code compilation, optimization, and performance. We encourage users to test and validate their applications in this new environment to ensure compatibility and performance.
Python Upgrades
The system Python has been upgraded to version 3.9, and the system Python 2 is no longer available on Ascend. Additionaly, newer versions of Python 3 are available through the modules system. This change may impact scripts and packages that rely on older versions of Python. We recommend users review and update their code to ensure compatibility or create custom environments as needed.
Available Software
Selected software packages have been installed on Ascend. You can use module spider to view the available packages after logging into Ascend. Additionally, check this page to see the available packages. Please note that the package list on the webpage is not yet complete.
After the Ascend cluster goes into full production (tentatively on March 31), you can view the installed software by visiting Browse Software and select "Ascend" under the "System".
If the software required for your research is not available, please contact OSC Help to reqeust the software.
Key change
A key change is that you are now required to specify the module version when loading any modules. For example, instead of using module load intel, you must use module load intel/2021.10.0. Failure to specify the version will result in an error message.
Below is an example message when loading gcc without specifying the version:
$ module load gcc Lmod has detected the following error: These module(s) or extension(s) exist but cannot be loaded as requested: "gcc". You encountered this error for one of the following reasons: 1. Missing version specification: On Ascend, you must specify an available version. 2. Missing required modules: Ensure you have loaded the appropriate compiler and MPI modules. Try: "module spider gcc" to view available versions or required modules. If you need further assistance, please contact oschelp@osc.edu with the subject line "lmod error: gcc"
Revised Software Modules
Some modules have been updated, renamed, or removed to align with the standards of the package managent system. For more details, please refer to the software page of the specific software you are interested in. Notable changes include:
| Package | Pitzer | Original Ascend | Ascend |
|---|---|---|---|
| Default MPI | mvapich2/2.3.3 | mvapich/2.3.7 | mvapich/3.0 |
| GCC | gnu | gnu | gcc |
| Intel MKL | intel, mkl | intel, mkl | intel-oneapi-mkl |
| Intel VTune | intel | intel | intel-oneapi-vtune |
| Intel TBB | intel | intel | intel-oneapi-tbb |
| Intel MPI | intelmpi | intelmpi | intel-oneapi-mpi |
| NetCDF | netcdf | netcdf-c, netcdf-cxx4, netcdf-fortran | |
| BLAST+ | blast | blast-plus | |
| Java | java | openjdk | |
| Quantum Espresso | espresso | quantum-espresso |
Licensed Software
No licensed software packages are available on Ascend.
Known Issues
We are actively identifying and addressing issues in the new environment. Please report any problems to the support team by contacting OSC Help to ensure a smooth transition. Notable issues include:
| Software | Versions | Issues |
|---|---|---|
Additional known issues can be found on our Known Issues page. To view issues related to the Ascend cluster, select "Ascend" under the "Category".
Batch Limit Rules
Memory limit
It is strongly suggested to consider the memory use to the available per-core memory when users request OSC resources for their jobs.
Summary
| Partition | # of gpus per node | Usable cores per node | default memory per core | max usable memory per node |
|---|---|---|---|---|
| nextgen | 2 | 120 | 4,027 MB | 471.91 GB |
| quad | 4 | 88 | 10,724 MB | 921.59 GB |
| batch | 4 | 88 | 10,724 MB | 921.59 GB |
It is recommended to let the default memory apply unless more control over memory is needed.
Note that if an entire node is requested, then the job is automatically granted the entire node's main memory. On the other hand, if a partial node is requested, then memory is granted based on the default memory per core.
See a more detailed explanation below.
Default memory limits
A job can request resources and allow the default memory to apply. If a job requires 300 GB for example:
#SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=30
This requests 30 cores, and each core will automatically be allocated 10.4 GB of memory for a quad GPU node (30 core * 10 GB memory = 300 GB memory).
Explicit memory requests
If needed, an explicit memory request can be added:
#SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=4 #SBATCH --mem=300G
See Job and storage charging for details.
CPU only jobs
We reserve 1 core per 1 GPU. The CPU-only job can be scheduled but can only request up to 118 cores per dual GPU node and up to 84 cores per quad GPU node. You can also request multiple nodes for one CPU-only job.
GPU Jobs
Jobs may request only parts of GPU node. These jobs may request up to the total cores on the node (88 cores) for quad GPU nodes.
Requests two gpus for one task:
#SBATCH --time=5:00:00 #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=20 #SBATCH --gpus-per-task=2
Requests two GPUs, one for each task:
#SBATCH --time=5:00:00 #SBATCH --nodes=1 #SBATCH --ntasks-per-node=2 #SBATCH --cpus-per-task=10 #SBATCH --gpus-per-task=1
Of course, jobs can request all the GPUs of a dense GPU node as well. These jobs have access to all cores as well.
Request an entire dense GPU node:
#SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=88 #SBATCH --gpus-per-node=4
Partition time and job size limits
Here is the walltime and node limits per job for different queues/partitions available on Ascend:
| Partition | Max walltime limit | Min job size | Max job size | Note |
|---|---|---|---|---|
| nextgen | 7-00:00:00 (168 hours) | 1 core | 16 nodes |
Can request multiple partial nodes For jobs requesting gpu=1 or 2 per node |
| quad | 7-00:00:00 (168 hours) | 1 core | 4 nodes |
Can request multiple partial nodes For jobs requesting gpu=3 or 4 per node |
| debug-nextgen | 1 hour | 1 core | 2 nodes | |
| debug-quad | 1 hour | 1 core | 2 nodes |
|
If you request -gpus-per-node=1 or -gpus-per-node=2 but need 80GB GPU memory node, please add --partition=quad
Usually, you do not need to specify the partition for a job and the scheduler will assign the right partition based on the requested resources. To specify a partition for a job, either add the flag --partition=<partition-name> to the sbatch command at submission time or add this line to the job script:
#SBATCH --paritition=<partition-name>
Job/Core Limits
| Max # of cores in use | Max # of GPUs in use | Max # of running jobs | Max # of jobs to submit | |
|---|---|---|---|---|
| Per user | 5,632 | 96 | 256 | 1000 |
| Per project | 5,632 | 96 | 512 | n/a |
An individual user can have up to the max concurrently running jobs and/or up to the max processors/cores in use. However, among all the users in a particular group/project, they can have up to the max concurrently running jobs and/or up to the max processors/cores in use.
Citation
For more information about citations of OSC, visit https://www.osc.edu/citation.
To cite Ascend, please use the following information:
Ohio Supercomputer Center. (2022). Ascend Cluster. Ohio Supercomputer Center. https://doi.org/10.82404/6JBT-FA57
BibTeX:
@MISC{Ohio_Supercomputer_Center2022-dl,
title = "Ascend Cluster",
author = "{Ohio Supercomputer Center}",
publisher = "Ohio Supercomputer Center",
year = "2022",
doi = "10.82404/6JBT-FA57"
}
ris:
TY - MISC AU - Ohio Supercomputer Center TI - Ascend Cluster PY - 2022 DA - 2022 PB - Ohio Supercomputer Center DO - 10.82404/6JBT-FA57 UR - http://dx.doi.org/10.82404/6JBT-FA57
Available software list on Next Gen Ascend
Available Software
- R: R/4.4.0
- afni: afni/2024.10.14
- alphafold: alphafold/2.3.2
- alphafold3: alphafold3/3.0.1
- amber: amber/24
- amd-hpc-benchmarks: amd-hpc-benchmarks/2024-10
- aocc: aocc/4.2.0, aocc/5.0.0
- app_code_server: app_code_server/4.8.3
- app_jupyter: app_jupyter/4.1.5
- bcftools: bcftools/1.17, bcftools/1.21
- bedtools2: bedtools2/2.31.0
- blast-database: blast-database/2024-07
- blast-plus: blast-plus/2.16.0
- blender: blender/4.2
- boost: boost/1.83.0
- bowtie: bowtie/1.3.1
- bowtie2: bowtie2/2.5.1
- bwa: bwa/0.7.17
- cmake: cmake/3.25.2
- connectome-workbench: connectome-workbench/1.3.2, connectome-workbench/2.0.0
- cp2k: cp2k/2023.2
- cuda: cuda/11.8.0, cuda/12.4.1, cuda/12.6.2
- cuda-samples: cuda-samples/11.8, cuda-samples/12.4.1, cuda-samples/12.6
- cudnn: cudnn/8.9.7.29-12
- cufflinks: cufflinks/2.2.1
- curl: curl/8.4.0
- darshan-runtime: darshan-runtime/3.4.6
- darshan-util: darshan-util/3.4.6
- dcm2nii: dcm2nii/11_12_2024
- desmond: desmond/2023.4
- dsi-studio: dsi-studio/2025.Jan
- fastqc: fastqc/0.12.1
- ffmpeg: ffmpeg/4.3.2, ffmpeg/6.1.1
- fftw: fftw/3.3.10
- fmriprep: fmriprep/20.2.0, fmriprep/24.1.1
- freesurfer: freesurfer/6.0.0, freesurfer/7.2.0, freesurfer/7.3.2, freesurfer/7.4.1
- fsl: fsl/6.0.7.13
- gatk: gatk/4.6.0.0
- gaussian: gaussian/g16c02
- gcc: gcc/12.3.0, gcc/13.2.0
- gdal: gdal/3.7.3
- geos: geos/3.12.0
- gromacs: gromacs/2024.4
- gsl: gsl/2.7.1
- gurobi: gurobi/12.0.0
- hdf5: hdf5/1.14.3
- hpctoolkit: hpctoolkit/2023.08.1
- hpcx: hpcx/2.17.1
- htslib: htslib/1.20
- intel: intel/2021.10.0
- intel-oneapi-mkl: intel-oneapi-mkl/2023.2.0, intel-oneapi-mkl/2024.1.0
- intel-oneapi-mpi: intel-oneapi-mpi/2021.10.0, intel-oneapi-mpi/2021.12.1
- intel-oneapi-tbb: intel-oneapi-tbb/2021.10.0
- intel-oneapi-vtune: intel-oneapi-vtune/2024.2.1
- julia: julia/1.10.4
- lammps: lammps/20230802.3
- libjpeg-turbo: libjpeg-turbo/3.0.2
- lightdesktop_base: lightdesktop_base/rhel9
- matlab: matlab/r2024a
- miniconda3: miniconda3/24.1.2-py310
- modules: modules/sp2025
- mricrogl: mricrogl/1.2.20220720
- mriqc: mriqc/0.16.1, mriqc/23.1.0rc0, mriqc/24.1.0
- mvapich: mvapich/3.0
- mvapich-plus: mvapich-plus/4.0
- mvapich2: mvapich2/2.3.7-1
- namd: namd/3.0
- nccl: nccl/2.19.3-1
- ncview: ncview/2.1.10
- netcdf-c: netcdf-c/4.8.1
- netcdf-cxx4: netcdf-cxx4/4.3.1
- netcdf-fortran: netcdf-fortran/4.6.1
- neuropointillist: neuropointillist/0.0.0.9000
- nextflow: nextflow/24.10.4
- node-js: node-js/20.12.0, node-js/22.12.0
- novnc: novnc/1.4.0
- nvhpc: nvhpc/24.11, nvhpc/25.1
- oneapi: oneapi/2023.2.3, oneapi/2024.1.0
- openfoam: openfoam/2312
- openjdk: openjdk/17.0.8.1_1
- openmpi: openmpi/5.0.2
- openmpi-cuda: openmpi-cuda/5.0.2
- orca: orca/5.0.4
- osu-micro-benchmarks: osu-micro-benchmarks/7.3
- parallel-netcdf: parallel-netcdf/1.12.3
- picard: picard/3.0.0
- proj: proj/9.2.1
- project: project/flowbelow, project/ondemand, project/pas1531
- python: python/3.12
- pytorch: pytorch/2.5.0
- qchem: qchem/6.2.1, qchem/6.2.2
- quantum-espresso: quantum-espresso/7.3.1
- reframe: reframe/3.11.2
- relion: relion/5.0.0
- rosetta: rosetta/3.12
- ruby: ruby/3.3.6
- samtools: samtools/1.17, samtools/1.21
- scipion: scipion/3.7.1
- snpeff: snpeff/5.2c
- spack: spack/0.21.1
- spark: spark/3.5.1
- spm: spm/8, spm/12.7771
- sratoolkit: sratoolkit/3.0.2
- star: star/2.7.10b
- texlive: texlive/2024
- topaz: topaz/anacondaApril25
- turbovnc: turbovnc/3.1.1
- vcftools: vcftools/0.1.16
- virtualgl: virtualgl/3.1.1
- visit: visit/3.3.3, visit/3.4.2
- xalt: xalt/latest
OSU College of Medicine Compute Service
Key information:
- To verify whether your project falls under CoM and can run jobs on Ascend only, run the following command from your terminal (on Cardinal or Ascend; On Pitzer, you’ll need to load Python first:
module load python/3.9-2022.05), replacing project_code with your actual project account:python /users/PZS0645/support/bin/parentCharge.py project_code - When using CoM project, please add
--partition=nextgenin your job scripts or specify ‘nextgen’ as the partition name with OnDemand apps. Failure to do so will result in your job being rejected. - A list of software available on Ascend can be found here: https://www.osc.edu/content/available_software_list_on_next_gen_ascend
- Always specify the module version when loading software. For example, instead of using module load intel, you must use
module load intel/2021.10.0. Failure to specify the version will result in an error message.
Hardware information:
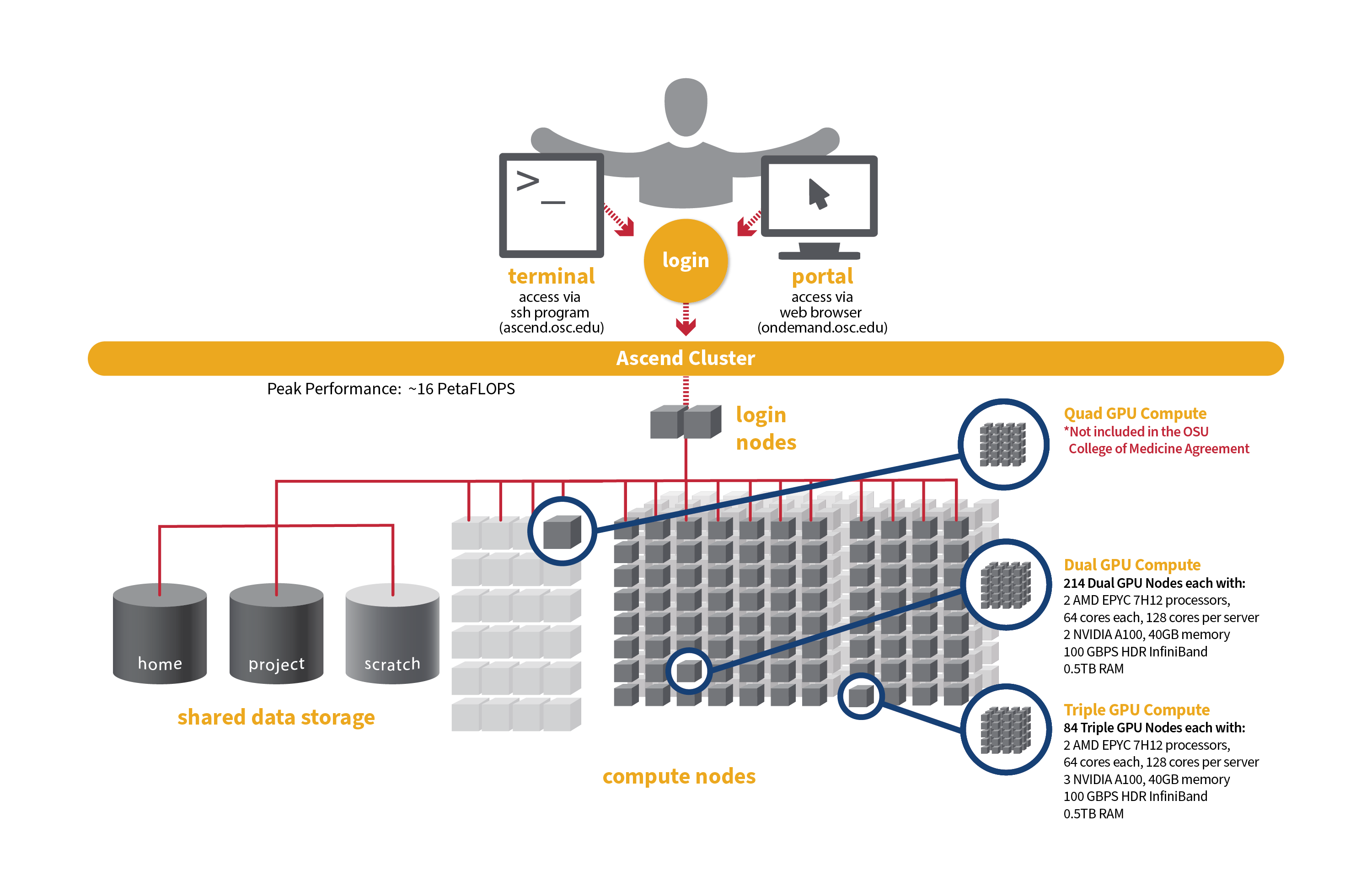
Detailed system specifications for Slurm workload:
- Dual GPU Compute: 190 Dell PowerEdge R7545 two-socket/dual GPU servers, each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 2 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
- Triple GPU Compute: 84 Dell PowerEdge R7545 two-socket/dual GPU servers, each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 3 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W (3rd GPU on each node is under testing and not available for user jobs)
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
Please check this Ascend page for more information on its hardware, programming and software environment, etc.
Governance
The CoM compute service is available to approved CoM users. A regular billing summary for all CoM PIs will be submitted to the OSU CoM Research Computing and Infrastructure Subcommittee (RISST) for review. PIs who are not eligible may be transitioned to a different agreement with OSC.
The committee will also review and consider requests for new project approvals or increases in storage quotas for existing projects.
Storage for CoM projects is billed to CoM at $3.20 per TB/month, with CoM covering up to 10TB. Any additional storage costs may be passed on to the PI.
Set up FY26 budgets
FY26 is the period of July 1, 2025 through June 30, 2026. As a reminder, the project budgets can only be managed by the project PI or a project administrator designated by the PI.
Do the following to create your budget for each project you want to use in FY26:
- Log into MyOSC
- Open the project details
- Select "Create a new budget"
- Select "Add or replace the CURRENT budget" to set the FY26 budget. Use 'unlimited' as the Budget type by choosing 'No' to the question: Do you want to set a dollar budget?
- Confirm your budget dates on the budget review page before submitting
- You will receive an email that your application has been submitted
It may be helpful to review a video explaining how to create and manage budgets.
Creating a new CoM project
Any user with the Primary Investigator (PI) role can request a new project in the client portal. Using the navigation bar, select Project, Create a new project. Fill in the required information.
If you are creating a new academic project
Choose ‘academic’ type as project type. Choose an existing charge account of yours in the College of Medicine, or if you do not have one, create a new charge account and select the department the work will be under. If you cannot find your department, please reach out to us for assistance. Use 'unlimited' as the Budget type by choosing 'No' to the question: Do you want to set a dollar budget?
For more instructions. see Video Tutorial and Projects, budgets and charge accounts page.
If you are creating a new classroom project
Choose ‘classroom’ type as project type. Under the top charge account of CoM: 34754, choose an existing charge account of yours, or if you do not have one, create a new charge account. You will request a $500 budget.
For more instructions. see Video Tutorial and Classroom Project Resource Guide.
Connecting
To access compute resources, you need to log in to Ascend at OSC by connecting to the following hostname:
ascend.osc.edu
You can either use an ssh client application or execute ssh on the command line in a terminal window as follows:
ssh <username>@ascend.osc.edu
From there, you can run programs interactively (only for small and test jobs) or through batch requests.
Running Jobs
OSC clusters are utilizing Slurm for job scheduling and resource management. Slurm , which stands for Simple Linux Utility for Resource Management, is a widely used open-source HPC resource management and scheduling system that originated at Lawrence Livermore National Laboratory. Please refer to this page for instructions on how to prepare and submit Slurm job scripts.
Remember to specify your project codes in the Slurm batch jobs, such that:
#SBATCH --account=PCON0000
where PCON0000 specifies your individual project code.
File Systems
CoM dedicated compute uses the same OSC mass storage environment as our other clusters. Large amounts of project storage is available on our Project storage service. Full details of the storage environment are available in our storage environment guide.
Training and Education Resources
The following are resource guides and select training materials available to OSC users:
- Users new to OSC are encouraged to refer to our New User Resource Guide page and an Introduction to OSC training video.
- A guide to the OSC Client Portal: MyOSC. MySC portal is primarily used for managing users on a project code, such as adding and/or removing users.
- Documentation on using OnDemand web portal can be found here.
- Training materials and tutorial on Unix Basics are here.
- Documentation on the use of the XDMoD tool for viewing job performance can be found here.
- The HOWTO pages, highlighting common activities users perform on our systems, are here.
- A guide on batch processing at OSC is here.
- For specific information about modules and file storage, please see the Batch Execution Environment page.
- Information on Pitzer programming environment can be found here.
Getting Support
Contact OSC Help if you have any other questions, or need other assistance.
SSH key fingerprints
- These are the public key fingerprints for Ascend:
ascend: ssh_host_rsa_key.pub = 2f:ad:ee:99:5a:f4:7f:0d:58:8f:d1:70:9d:e4:f4:16
ascend: ssh_host_ed25519_key.pub = 6b:0e:f1:fb:10:da:8c:0b:36:12:04:57:2b:2c:2b:4d
ascend: ssh_host_ecdsa_key.pub = f4:6f:b5:d2:fa:96:02:73:9a:40:5e:cf:ad:6d:19:e5
These are the SHA256 hashes:
ascend: ssh_host_rsa_key.pub = SHA256:4l25PJOI9sDUaz9NjUJ9z/GIiw0QV/h86DOoudzk4oQ
ascend: ssh_host_ed25519_key.pub = SHA256:pvz/XrtS+PPv4nsn6G10Nfc7yM7CtWoTnkgQwz+WmNY
ascend: ssh_host_ecdsa_key.pub = SHA256:giMUelxDSD8BTWwyECO10SCohi3ahLPBtkL2qJ3l080
Technical Specifications
The following are technical specifications for Quad GPU nodes.
- Number of Nodes
-
24 nodes
- Number of CPU Sockets
-
48 (2 sockets/node)
- Number of CPU Cores
-
2,304 (96 cores/node)
- Cores Per Node
-
96 cores/node (88 usable cores/node)
- Internal Storage
-
12.8 TB NVMe internal storage
- Compute CPU Specifications
-
AMD EPYC 7643 (Milan) processors for compute
- 2.3 GHz
- 48 cores per processor
- Computer Server Specifications
-
24 Dell XE8545 servers
- Accelerator Specifications
-
4 NVIDIA A100 GPUs with 80GB memory each, supercharged by NVIDIA NVLink
- Number of Accelerator Nodes
-
24 total
- Total Memory
- ~ 24 TB
- Physical Memory Per Node
-
1 TB
- Physical Memory Per Core
-
10.6 GB
- Interconnect
-
Mellanox/NVIDA 200 Gbps HDR InfiniBand
-
The following are technical specifications for Triple GPU nodes.
- Number of Nodes
-
84 nodes
- Number of CPU Sockets
-
168 (2 sockets/node)
- Number of CPU Cores
-
10,752 (128 cores/node)
- Cores Per Node
-
128 cores/node (120 usable cores/node)
- Internal Storage
-
1.92 TB NVMe internal storage
- Compute CPU Specifications
-
2 AMD EPYC 7H12 processors for compute
- 2.60 GHz
- 64 cores per processor
- Computer Server Specifications
-
84 Dell R7525 servers
- Accelerator Specifications
- 3 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W
- Number of Accelerator Nodes
-
168 total
- Total Memory
- ~ 42 TB
- Physical Memory Per Node
-
0.5 TB
- Physical Memory Per Core
-
4 GB
- Interconnect
-
HDR100 Infiniband (100 Gbps)
-
The following are technical specifications for Dual GPU nodes.
- Number of Nodes
-
214 nodes ( 190 nodes available to Slurm and 24 reserved for Kubernetes)
- Number of CPU Sockets
-
380 (2 sockets/node)
- Number of CPU Cores
-
24,320 (128 cores/node)
- Cores Per Node
-
128 cores/node (120 usable cores/node)
- Internal Storage
-
1.92 TB NVMe internal storage
- Compute CPU Specifications
-
2 AMD EPYC 7H12 processors for compute
- 2.60 GHz
- 64 cores per processor
- Computer Server Specifications
-
190 Dell R7525 servers
- Accelerator Specifications
- 2 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W
- Number of Accelerator Nodes
-
380 total
- Total Memory
- ~ 95 TB
- Physical Memory Per Node
-
0.5 TB
- Physical Memory Per Core
-
4 GB
- Interconnect
-
HDR100 Infiniband (100 Gbps)