2009 Research Highlights | Research Demos | Our SC09 Partners | SC09 Videos

Project lead:
Daniel Janies, Ph.D.,
The Ohio State University
Research title:
Unified genomic, geographic, and evolutionary approaches to understand the spread of infectious disease
Funding sources:
National Science Foundation, Defense Advanced Research Projects Agency, The Ohio State University
A biomedical informatics researcher at the Ohio State University Medical Center is using supercomputers to synthesize datasets of many genomes to help monitor the global spread of emergent infectious diseases.
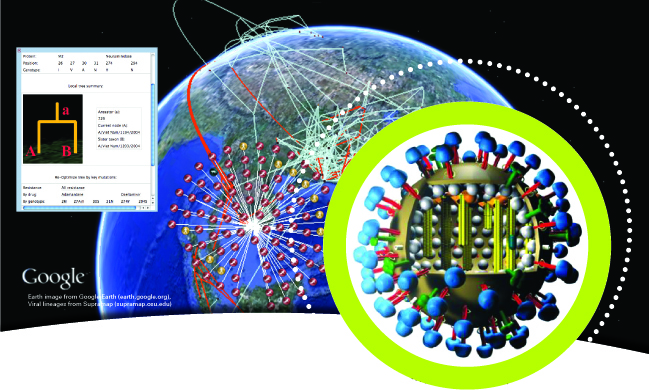
OSU associate professor Daniel Janies, an expert in computational genomics, uses sophisticated Ohio Supercomputer Center systems to calculate phylogenetic trees of the spread of influenza, such as H1N1 "swine flu" and H5N1 "avian flu," across various hosts, time and geography.
He then turns to an easyto- use web application, Supramap, he and his colleagues created in 2007 to map the spread of agents of infectious diseases (such as the H5N1 "avian flu" virus, shown above) into virtual globes such as Google Earth based on genomic and geographic data. The results are akin to weather maps for disease that allow public health officials to visualize when and where pathogens jump from animals to humans and evolve to resist drugs.
"Our ultimate goal is to help predict where the next outbreak of the virus is likely to occur," he said. "When we're done, we will be able to tell you where each viral gene came from in the world and what mutations are specific to the outbreak lineage. We then share these tools and maps so that diseases can be diagnosed and treated rapidly."
For more information, see: http://supramap.osu.edu