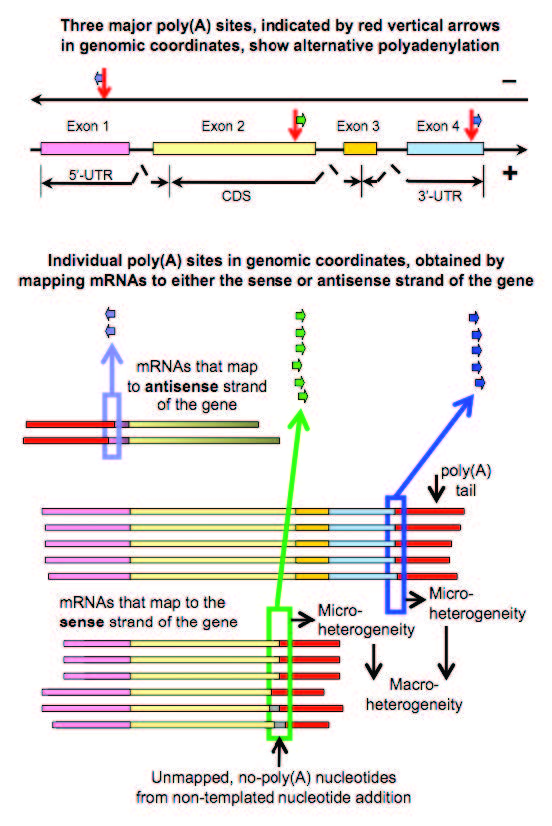
When eukaryotic genes are expressed, precursor messenger RNA (pre-mRNA) must first be processed to become mature mRNA. One step of the maturation process is constitutive polyadenylation: the attachment of a poly(A) tail to mark and protect the end of mRNA. However, alternative polyadenylation at a different poly(A) sites on the pre-mRNA might result in information loss and sometimes can cause cancers and other diseases in humans.
A Miami University research team led by Chun Liang, Ph.D., is investigating potential regulatory mechanisms controlling alternative and constitutive polyadenylation within eight different species – two diatoms, two green algae, spikemoss, moss, Arabidopsis and human.
“It has been shown that polyadenylation is guided by regulatory elements, or motifs, known as the poly(A) signals,” said Liang, an assistant professor in bioinformatics. “The process is carried out by the protein complex that recognizes and binds to those motifs, cleaves the mRNA and conducts polyadenylation.”
Classical genetic analysis and recent genome-wide bioinformatics analysis suggest that there are three typical regulatory elements in plants: a cleavage element, a near-upstream element and a far-upstream element.
“Using the powerful bioinformatics software package MEME (Multiple EM for Motif Elicitation), we are searching for over-represented sequence motifs from each of the regions,” Liang explained. “Our preliminary data analysis reveals interesting differences among different species, but the real challenge is that we have to process a large amount of data to detect significant/meaningful patterns, while processing a large amount of data demands more computational resources.”
--
Project lead: Chun Liang, Miami University
Research title: Comparative analysis of polyadenylation signals in eukaryotes
Funding source: Miami University