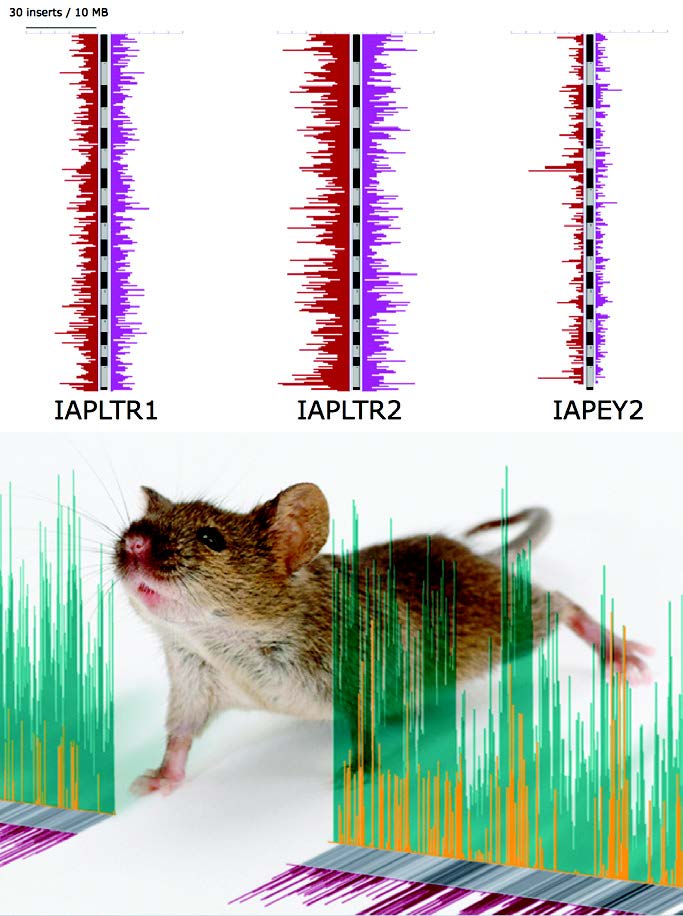
Recently, large-scale sequencing projects have described the complete DNA blueprints for humans, the mouse and other organisms. This work highlighted the surprising fact that almost half of all mammalian DNA genomes is made up of mobile genetic elements called retrotransposons. These virus-like retrotransposons, sometimes called “jumping genes,” can move from one chromosomal location to another, causing variation between individuals or lineages and occasional genetic disruptions resulting in diseases including cancers.
The controls and consequences of movement by these mobile elements are a focus of research for David Symer, M.D., Ph.D., and Keiko Akagi, Ph.D., in the Department of Molecular Virology, Immunology and Medical Genetics and the Human Cancer Genetics Program at The Ohio State University Comprehensive Cancer Center. Symer and Akagi access OSC’s Glenn Cluster to run various genomic alignment programs to align hundreds of millions of short sequence genomic traces – massive datasets generated from previously unsequenced individuals or cells – against the enormous reference genome assemblies for human, mouse and other organisms. They also utilize RepeatMasker and other data-intensive bioinformatics programs.
“Our long-term goal is to understand the biological and evolutionary roles played by transposons. These ‘jumping genes’ are major drivers of genomic variation. They can affect gene expression and structure, chromatinization and chromosomal structures,” said Symer. “We’d like to exploit practical applications of this new knowledge, to improve diagnosis and treatment of human cancers and other diseases, and by modeling such processes in other organisms.”
--
Project lead: David E. Symer, The Ohio State University
Research title: Identification and analysis of genomic polymorphisms that distinguish mouse and human lineages and populations
Funding sources: National Cancer Institute (National Institutes of Health); The Ohio State University