above: Ohio State’s Rebecca Lamb analyzed the evolutionary history of the poly(ADP-ribose)polymerase (PARP) enzyme superfamily, a protein group implicated in a wide range of human diseases and, therefore, serve as important targets for anti-cancer therapies.
A group of enzymes implicated in a wide range of human diseases are important targets for anti-cancer therapies and the targets of a study by a molecular biologist to more fully understand the enzymes by helping to better define the group's family tree.
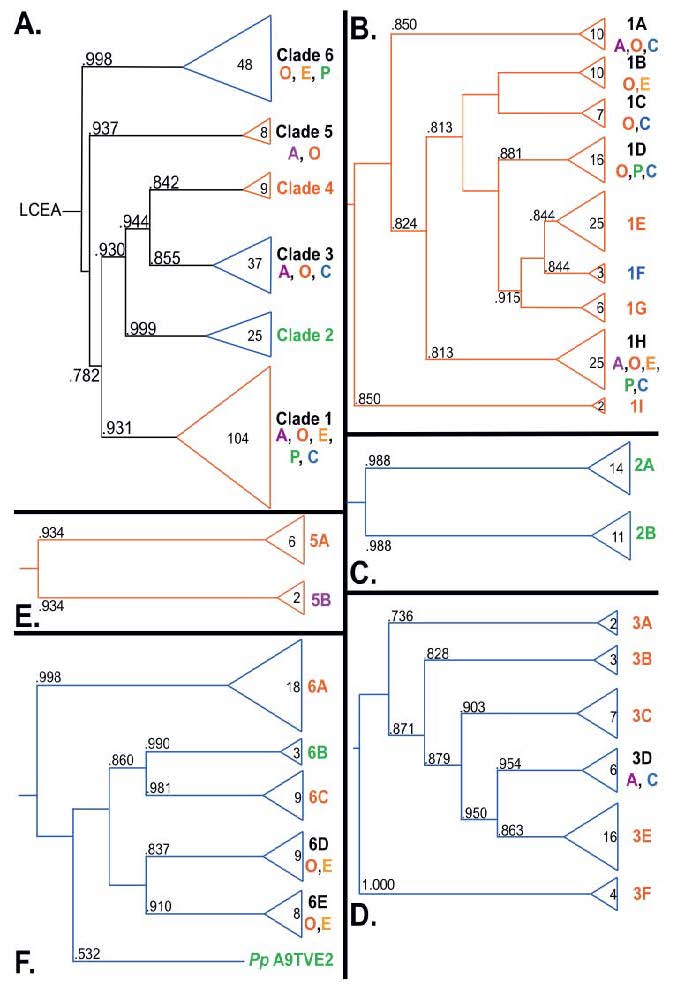
Along with several of her Ohio State University colleagues, Rebecca S. Lamb, Ph.D., an assistant professor of molecular genetics, recently analyzed the evolutionary history of the poly(ADP-ribose)polymerase (PARP) superfamily. These proteins are found in eukaryotes, a wide range of organisms – animals, plants, molds, fungi, algae and protozoa – whose cells contain complex structures enclosed within membranes. While PARP proteins can be found within any of these 'supergroups,' they have been studied most extensively in mammals.
“In these organisms, PARPs have key functions in DNA repair, genome integrity and epigenetic regulation,” said Lamb. “More recently it has been found that proteins within the PARP superfamily have a broader range of functions than initially predicted.”
The researchers used computers to identify 236 PARP proteins from 77 species across five of the six supergroups. Lamb then accessed systems at the Ohio Supercomputer Center to perform extensive phylogenetic analyses of the identified PARP regions, computationally intensive work that would have been impossible without such resources, according to Lamb. In particular, the ability to try a variety of tools that require a great deal of CPU and memory capabilities was essential. Among the software tools she employed was the PhyML3.0 package, which fit a statistical model to the aligned sequence data and provided estimates for the model’s parameters.
“PARPs are found in all eukaryotic supergroups for which sequences are available, but some individual lineages within supergroups have independently lost these genes,” said Lamb. “The PARP superfamily can be subdivided into six branches or ‘clades.’ Two of these clades were likely found in the last common eukaryotic ancestor. In addition, we have identified PARPs in organisms for which they have not previously been described.”
Three main conclusions were drawn from the study. First, the broad distribution and pattern of representation of PARP genes indicated to the researchers that the ancestor of all existing eukaryotes encoded proteins of this type and may have had multiple members. Second, the ancestral PARP proteins had different functions and activities. One of these proteins likely functioned in DNA damage response. Third, the diversity of the PARP superfamily is larger than previously documented, suggesting as more eukaryotic genomes become available, this gene family will grow in both number and type.
--
Project lead: Rebecca S. Lamb, The Ohio State University
Research title: Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes
Funding sources: Ohio Plant Biotechnology Consortium, The Ohio State University
Web site: www.biosci.ohio-state.edu/pcmb/osu_pcmb/faculty_sites/rebecca_lamb/index...