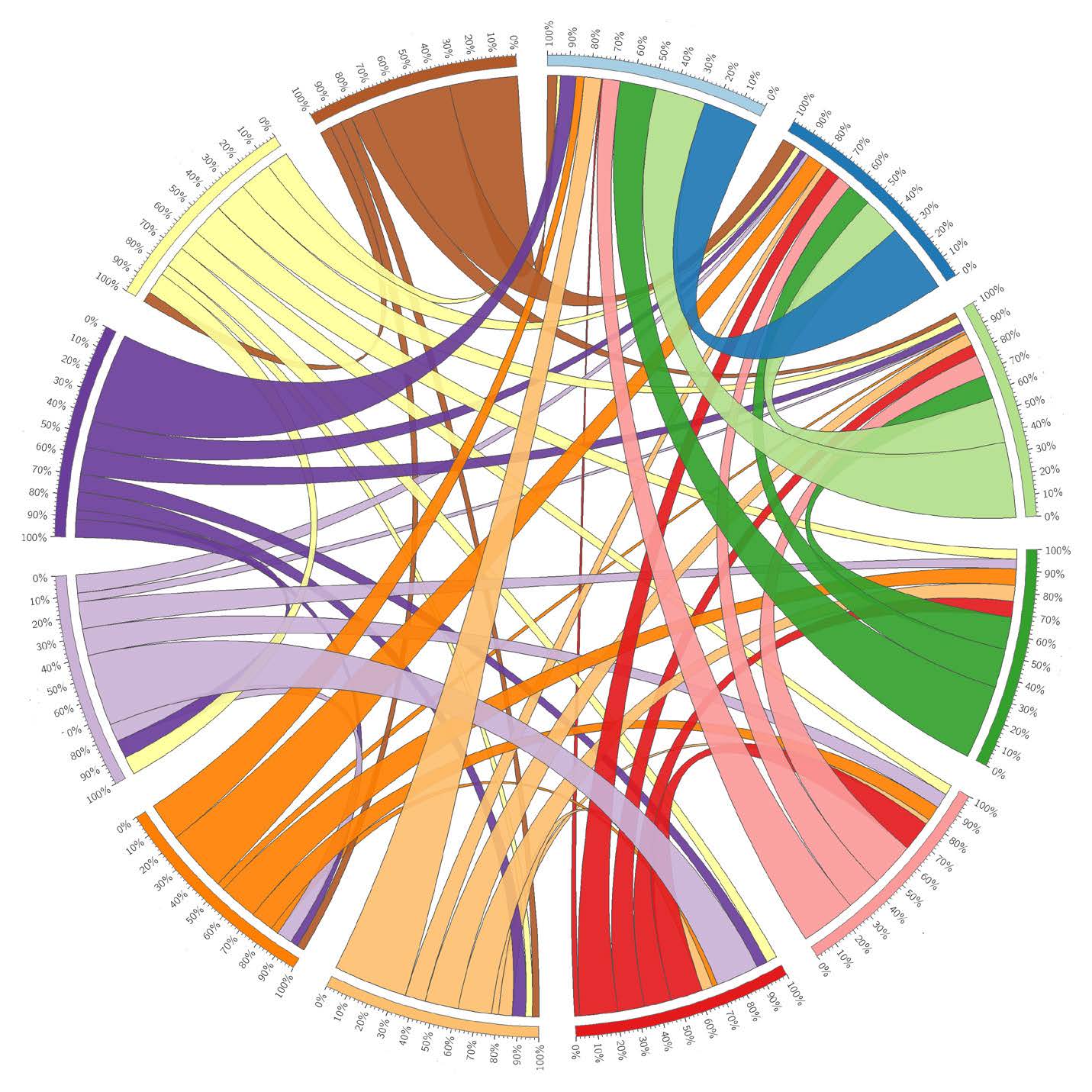
Distilled from a 134-patient study, this Circos plot links differentially methylated regions shared between mutation subgroups.
At The Ohio State University Comprehensive Cancer Center – Arthur G. James Cancer Hospital and Richard J. Solove Research Institute (OSUCCC), cancer researchers turn to the multifunctional Nucleic Acid Shared Resource (NASR) Illumina Core to analyze genomic and epigenomic influences on the disease – and indirectly, the Ohio Supercomputer Center.
“We provide reliable, high-quality, affordable, computation support for researchers who sequence epigenomics libraries in our Core, whether they are from The James, The Ohio State University, The Ohio State University Wexner Medical Center, or another university,” said Pearlly Yan, technical director, NASR Illumina Core.
Specifically, NASR provides the research community with centralized instrumentation and expertise for Sanger-based and non-Sanger-based DNA sequencing, genotyping, DNA methylation analysis and gene expression analysis. It also offers accessory equipment for quantitative measurement and quality control of nucleic acids, and nucleic acid imaging.
“Next-generation sequencing files typically generate several Gigabits of data per sample and datasets often exceed 100 samples,” Yan said. “Analyzing such large datasets requires computationally intensive, complex calculations for such processes as quality control after sequencing, genomic alignment, and custom downstream analyses.”
The primary vehicle for this analysis is the lllumina HiSeq 2000, a machine specifically designed for sequencing genomes, methylomes and transcriptomes that allows researchers to quickly sequence large volumes of samples. For example, with the current data output, up to 192 indexed samples can be sequenced per run (two concurrent HiSeq flow cells generating 20 million reads per sample) for high-throughput gene expression profiling study. If the intent is to perform high-resolution transcriptome analysis, more than 50 samples can be analyzed per run at about 100 million reads per sample.
“The collaboration between the Ohio Supercomputer Center and our Core is invaluable in turning large amounts of sequencing data into information that drives clinical and translational research in OSUCCC and Ohio State’s Wexner Medical Center. This type of collaboration has allowed us to provide better services to our users and timely custom analyses of sequencing data,” Yan said. “Also, a good portion of our computation staff members are undergraduate assistants. Their ability to perform data analysis in the OSC environment offers them the unique education experience of learning about both cutting edge genomic data and the supercomputer environment.”
The shared resources at Ohio State’s Comprehensive Cancer Center – James Cancer Hospital and Solove Research Institute are an NCI-recognized network of specialized service facilities, or core facilities, which facilitate an investigator’s ability to conduct cancer research by reaching across medical disciplines.
--
Project lead: Pearlly Yan, The Ohio State University
Research title: OSU Comprehensive Cancer Center NASR-Illumina core epigenetics projects
Funding source: The Ohio State University
Web site: http://bit.ly/OSC-RR-Yan