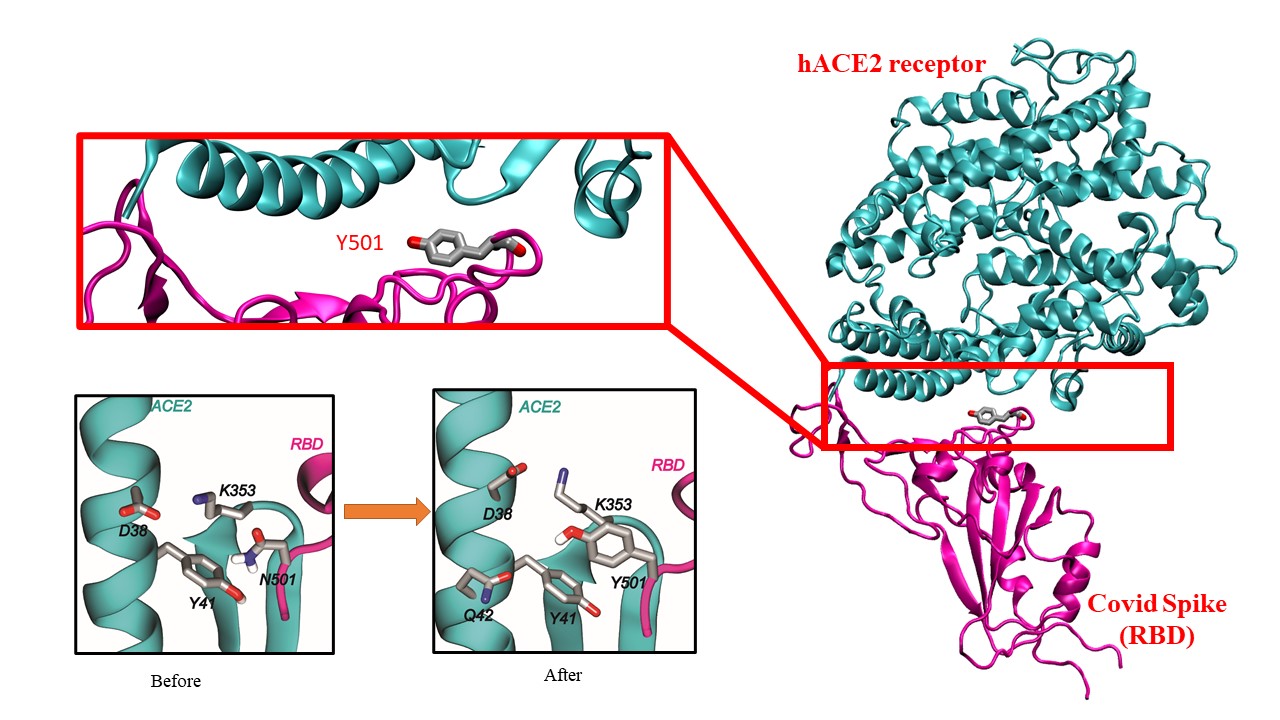
Right panel: A figure displaying the binding of the receptor binding domain (RBD, colored in magenta) of the mutated SARS-CoV2 spike protein with the human cell surface receptor ACE2 (hACE2, colored in cyan). Lower left panel: the binding interactions for the 501th residue of SARs-CoV2 before and after the N501Y mutation.
With the COVID-19 pandemic presenting an ongoing global challenge, Xiche Hu’s lab at the University of Toledo is taking a closer look at the mutations of the coronavirus.
For more than 20 years, Hu’s lab has been researching an issue known as molecular recognition, which is how two molecules locate each other and bind together to perform a biological function. When the COVID-19 pandemic started, Hu wondered if his molecular recognition expertise could help scientists understand how the coronavirus identifies the right receptors to bind to in the human body to trigger infection.
“We immediately realized that there were many molecular recognition issues, so we felt duty bound to study this,” he said. “The Ohio Supercomputer Center (OSC) had called for researchers studying the coronavirus, offering a special grant. We applied and were granted priority access and an unlimited budget for 18 months."
Hu and two doctoral students at University of Toledo, Pawan Bhatta and Majed Aljohani, began by studying the atomic-level differences between the original SARS-CoV-2 and the new variants of concern.
Using high-level quantum mechanics calculations, Hu’s group can determine the binding energy between COVID-19’s spike proteins and the human receptors within the cell. A greater binding energy can make the virus more contagious.
Making an accurate calculation of binding energy requires highly complex computational algorithms and vast computational resources.
“If we did one of these calculations in our lab, we are talking about weeks or months, but when we do it at OSC we can finish it within days,” Hu said. “We often need to do multiple calculations in parallel, so the OSC resources are instrumental to this research.”
In four out of five variants of concern, researchers found that a common mutation, N501Y, increased the binding energy between parts of the human cell and the virus. In addition, the scientists discovered that the mutation enables the virus to form a network of water molecules that boosts the variant’s ability to infect cells.
These discoveries may allow the scientific community to further develop methods to fight the COVID-19 pandemic.
“With this research we can move to the next step, screening a library of compounds to find which molecules can block out this site, preventing infection,” Hu said.