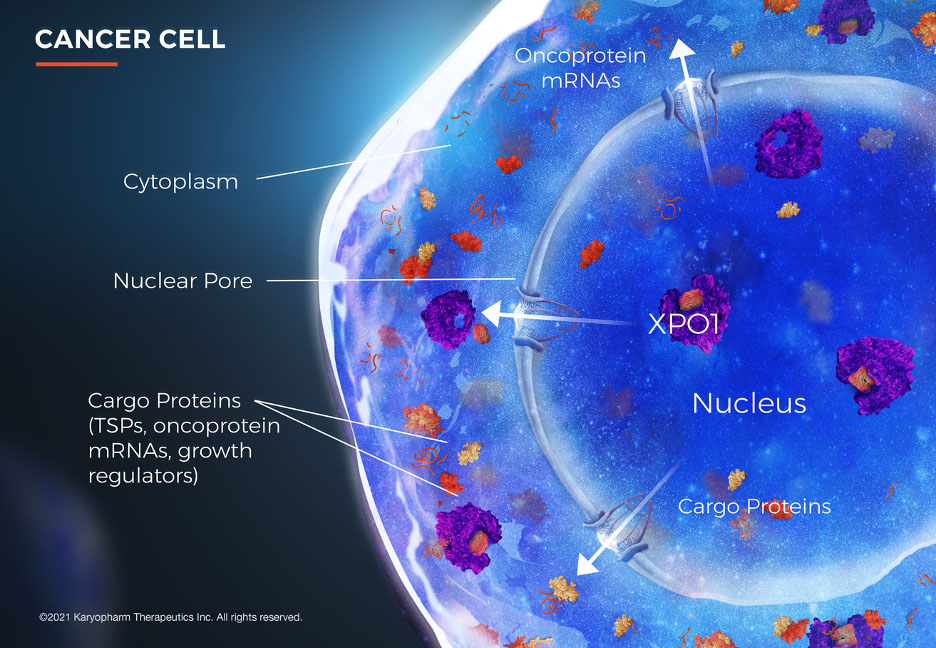
The karyopherin protein exportin 1 (XPO1) facilitates the shuttling of hundreds of its cargo proteins from the nucleus to the cytoplasm. XPO1 is over-expressed by many types of cancer cells. Image Copyright 2021 Karyopharm Therapeutics Inc. All Rights Reserved.
A drug designed to treat a certain type of cancerous tumor might work well in some patients but not others. To determine why, scientists can study whether specific genetic mutations may impact the therapy’s effectiveness.
Karyopharm Therapeutics, a commercial-stage pharmaceutical company pioneering novel cancer therapies, is taking a closer look at these unique molecular characteristics of different cancers with the help of the Ohio Supercomputer Center (OSC).
Karyopharm's lead selective inhibitor of nuclear export (SINE) compound and first-in-class, oral exportin 1 (XPO1) inhibitor, XPOVIO® (selinexor), is approved in the U.S. Selinexor is marketed by the company in three oncology indications and has received regulatory approvals in various indications in a growing number of ex-U.S. territories and countries, including Europe and the United Kingdom (as NEXPOVIO®), China and Singapore.
Christopher Walker, an alumnus of The Ohio State University, serves as the company’s director of bioinformatics. Walker conducts genetic sequencing of cancerous tumors and analyzes data to help Karyopharm predict potential responsiveness and whether or not selinexor may be effective in a certain population. The work requires high performance computing power and relevant software programs that can manage a large volume of raw DNA and RNA sequencing data.
During his time as a postdoctoral and research fellow at Ohio State, Walker conducted oncology research using OSC resources. OSC was a critical resource for his projects, which included targeted sequencing of the then-largest cohort of endometrioid endometrial cancer samples, RNA sequencing of acute myeloid leukemia samples and exome sequencing of thyroid cancer samples. After joining Karyopharm Therapeutics, Walker sought to establish a bioinformatics infrastructure for the company. He determined that OSC would be a valuable asset to support Karyopharm’s computational processing needs, and so chose to continue partnering with the Center in his new role. Working with OSC to enhance the pharmaceutical company’s bioinformatics capabilities has been preferable to trying to build or maintain such resources in-house, he said.
“The infrastructure is there—it’s set up to be really easy to use—and the software we use for processing next-generation sequencing data is preloaded and well-maintained at OSC,” Walker said. “It’s amazing to see how often OSC is as able to upgrade their available hardware. The addition of the Pitzer cluster was really impressive.”
The research partnership is yielding useful results for Karyopharm Therapeutics. In a published study of patients with advanced, refractory dedifferentiated liposarcoma, for which selinexor is being investigated, Walker and colleagues found that selinexor treatment may be effective against tumors that did not express a gene called CALB1. Additionally, at the 2022 American Society of Clinical Oncology conference, results from Karyopharm’s Phase III SIENDO study were presented, showing that TP53 mutation status may be associated with response to selinexor. Further investigation is needed.
Walker is excited about a new era in science in which bioinformatics may be used to predict the patients most likely to benefit from new therapies. “You’ll find more and more drugs being approved for specific malignancies with a specific mutation type and maybe even with a specific expression profile at the RNA level in the near future,” Walker said. “That’s what I envision in the field.”