For Sydney Decker, evolution isn’t just a topic from a biology textbook—it’s the foundation of her scientific curiosity. A postdoctoral researcher in the College of Arts and Sciences at The Ohio State University, Decker has been driven by one overarching question since the beginning of her undergraduate career nearly 10 years ago.
“I am really interested in how species evolve and how new species form,” Decker said. “That’s the big question that’s always motivated my work.”
That question led her to the world of yellow bats—a group of small, visually similar mammals found from the southern United States through Central and South America, down to Argentina. These tropical bats roost in palm trees rather than caves or buildings, earning them the nickname “palm tree dudes,” as Decker fondly calls them.

Because the bats look nearly identical but are spread across a wide geographic range, researchers suspected that they could be an example of how geographic isolation and ecological pressures can drive the formation of new species.
“They’re morphologically very similar,” Decker said, “but genetically, they can be quite distinct. That contrast is part of what makes them so fascinating.”
To explore those differences, Decker sequenced more than 40,000 genomic loci—specific locations on a chromosome—from 132 yellow bat specimens, many drawn from museum collections. She paired this dataset with high-resolution micro-computed tomography (micro-CT) scans of bat skulls, creating detailed 3D images of their morphology.
As she began generating and analyzing these massive datasets, Decker quickly realized she would need high performance computing to make her research possible. She turned to the Ohio Supercomputer Center (OSC), where she had already been introduced to supercomputing through a related project.
“I started using OSC right away,” she said. “The scale of the genomic data I collected is pretty massive. It’s really not possible to analyze on a personal laptop, or even really on the workstations that are available to me.”
With OSC’s resources, Decker was able to run large-scale analyses, test computationally demanding models, and store datasets that would otherwise be unmanageable.
“I really couldn’t do this work without it. It’s not just about speed—it’s about access. Having OSC means I can actually do this kind of research,” said Decker, who presented her findings at the 2025 OSC Research Symposium.
The genomic results gathered through OSC were striking: The yellow bats that were once thought to represent four species actually contain seven distinct lineages. The genetic divergence appears to date back to the Pleistocene epoch (often referred to as the ice age), when climatic shifts likely caused populations to separate and evolve independently. Discovering hidden biodiversity like this can reshape understanding of how species form, and it can influence conservation decisions by identifying distinct lineages that may need protection.
The discovery also raised a new question: If these lineages are genetically distinct, why don’t the bats’ bodies show clearer differences? To answer that, Decker returned to the micro-CT scans, comparing detailed 3D images of skulls to see whether subtle physical traits could help bridge the gap between what the DNA revealed and what morphology showed. While the 3D images helped distinguish some species, they failed to reveal differences in the new lineages uncovered through DNA. Researchers needed to reconcile the two datasets.
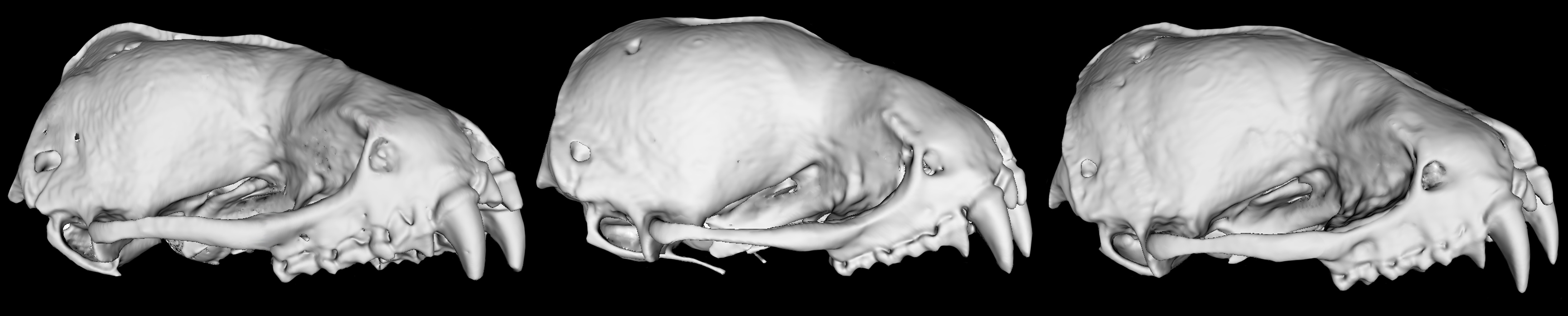
In collaboration with the Imageomics Institute in Ohio State’s College of Engineering, Decker is developing machine learning models that can extract far more information from CT scans than traditional approaches. By training algorithms to recognize patterns too subtle for the human eye, she hopes to identify morphological signals that align with the genetic differences.
“That will hopefully bridge the gap between what the morphology shows and what the genomics tell us,” Decker said.
Her work ties directly into the research program of her advisor, Bryan Carstens, an evolutionary biologist at Ohio State whose lab focuses on uncovering hidden biodiversity and developing new tools to improve species delimitation. Like Decker, Carstens also relies on OSC to power his lab’s large-scale genomic analyses.
Decker’s project pushes that work forward by integrating machine learning with traditional genomic and morphological approaches—helping to build a framework for biodiversity studies that is both more accurate and more comprehensive.
This integrative framework—combining genomics, morphology, and machine learning—could give evolutionary biologists a more accurate model of how species form and diverge. It also has broad potential applications for biodiversity studies, from refining taxonomy to improving conservation efforts by recognizing distinct lineages that need protection.
As she continues refining her models and drawing connections between morphology and genetics, Decker is also thinking about her future—one that, like evolution itself, is full of possibility and uncertainty.
“I think this kind of interdisciplinary work is where the field is headed,” she said. “And I’m really grateful to be part of that shift.”
Written by Lexi Biasi
The Ohio Supercomputer Center (OSC) addresses the rising computational demands of academic and industrial research communities by providing a robust shared infrastructure and proven expertise in advanced modeling, simulation and analysis. OSC empowers scientists with the services essential to making extraordinary discoveries and innovations, partners with businesses and industry to leverage computational science as a competitive force in the global knowledge economy and leads efforts to equip the workforce with the key technology skills required for 21st century jobs.