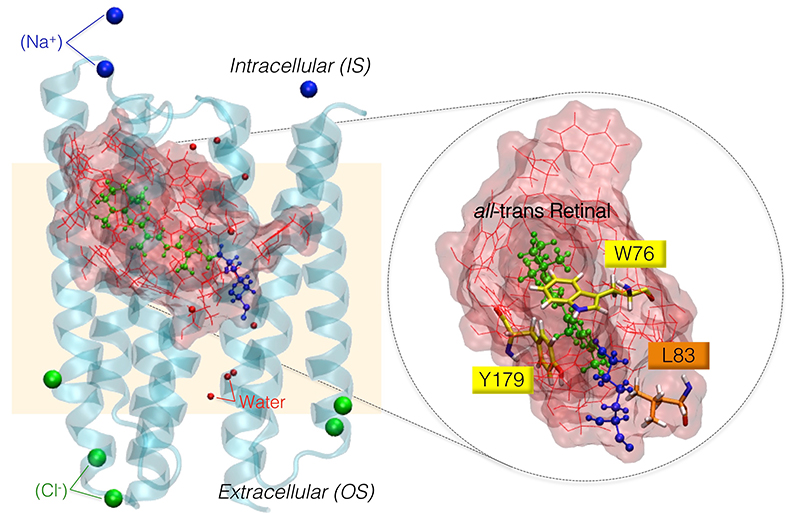
In the emerging field of optogenetics, scientists are working to develop light-responsive proteins (photoreceptors) that will allow them to observe the nerve impulses in the brain or to control specific cellular features, such as metabolic pathways, gene expression and ion channels.
Massimo Olivucci, Ph.D., research professor and director of the Laboratory for Computational Photochemistry and Photobiology at Bowling Green State University is leveraging Ohio Supercomputer Center (OSC) services to study the optogenetic potential of Anabaena sensory rhodopsin, a light-sensing protein found in a cyanobacterium, or blue-green algae. Rhodopsin proteins are present in almost every organism featuring “vision” in a basic or complex form.
“We are trying to understand the details of molecular mechanisms allowing biological organisms to capture and use light energy and how this knowledge could then be exploited to design new materials,” Olivucci said. “For this reason, we use computers to model the lighttriggered dynamics of biological photoreceptors at atomic-level resolution.”
Olivucci’s group is also modeling the dynamics of rhodopsin mutants—in principle, thousands of mutants—in order to find molecular modifications that will produce high levels of fluorescence. More specifically, the lab is screening mutant models to isolate a few promising variants that biologists then attempt to produce in the lab. In a sense, the group is attempting to simulate biological evolution under a selective pressure for maximizing fluorescence. To facilitate this computationally intensive process, Olivucci is leasing a small group of very powerful nodes on OSC’s Owens Cluster— what is known in the supercomputing industry as a “condo.”
“Given the large number of calculations we need to run, we need extremely fast processors,” Olivucci said. “Even more importantly, these are very long calculations, and the condo arrangement with OSC offers us the opportunity to run jobs for an entire week.”
Olivucci also points out that the number of variants they are screening is far beyond that which an individual is able to process manually in a reasonable timeframe. Therefore, his group is continuously working to enhance their Automatic Rhodopsin Modeling (ARM) software to build and evaluate thousands of computer models of variants.
Additionally, this research is providing students on his team with extremely valuable experience. Pharmaceutical companies today, for instance, design new drugs by simulating drug interactions within a computer model before investing enormous amounts of time and money testing them in the laboratory or on live subjects.
__________
PROJECT LEAD // Massimo Olivucci, Ph.D., Bowling Green State University
RESEARCH TITLE // Investigating Anabaena sensory rhodopsin mutants for the design of novel optogenetic tools
FUNDING SOURCE // National Science Foundation
WEBSITE // ccmaol1.chim.unisi.it