The cornerstone of an effective therapeutic drug development program is a rock-solid computational protocol that accurately and efficiently illustrates how molecules interact within the medicine and inside the human body. That information can be used to help fight and cure disease.
But it first requires someone to get deep into the weeds to study the physics of how druglike molecules bind together with molecular machines such as proteins. Manori Jayasinghe, Ph.D., an associate professor of physics at the University of Cincinnati Blue Ash College, is such a person.
Jayasinghe researches computational molecular biophysics and, as one might expect, that work is computationally intense. That is why Jayasinghe uses the world-class storage and high performance computing systems of the Ohio Supercomputer Center (OSC).
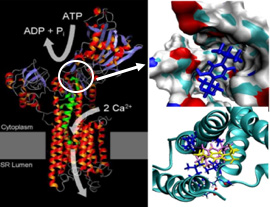
Jayasinghe’s current research involves modeling “the structure, dynamics and processes of biomolecular machines, like proteins or DNA,” as she works toward deep insight into how the subunits of “large biomolecular systems cooperate to work like a real machine to perform their intended action in the living cell.”
In order to develop an understanding of the structure, dynamics and processes of proteins and DNA, Jayasinghe performs numerous molecular dynamics simulations on the Oakley and Owens clusters at OSC. To accomplish that work, she relies on a variety of software packages, such as CHARMM, Python, Gromacs, Gaussian and MatLab.
“Parallel processing, such as the CHARMM program we use for our simulations, efficiently use high-bandwidth, low-latency, communication between cores to distribute computational tasks to multiple processors,” Jayasinghe said. “We can get results five, six times faster that way than with single processor computations.”
As those results get back to Jayasinghe, she can identify high-potency, drug-like compounds that bind with targeted proteins and help drug developers in their synthesis process. One recent project on which she teamed with OSC involved unraveling the reactions within proteins that are the target for anti-cancer medicines in prostate cancer therapy.
That information will help Jayasinghe develop computational methods to estimate the effectiveness of drug potency on specific proteins.
“That protocol will be used to study other inhibitor/enzyme systems in the future,” Jayasinghe said. “This knowledge at the molecular level and the inhibition mechanisms (which slow or prevent chemical reactions) will serve as a great resource of information to characterize mechanisms of enzyme inhibition in general.”
_____________
PROJECT LEAD // Manori Jayasinghe, Ph.D., University of Cincinnati Blue Ash College
RESEARCH TITLE // Evaluation of absolute binding free energies of BHQ (2,5 di-tert-butylhydroquinone) analogues binding of human Ca2+ ion transporter (SERCA)
FUNDING SOURCE // University of Cincinnati
WEBSITE // linkedin.com/in/manori-jayasinghe-945b9a32