Matthew Sullivan, Ph.D., and the Ohio Supercomputer Center (OSC) have teamed up to give scientists insight into how to better study viruses found in a variety of communities. This information could prove invaluable to understanding everything from what’s going on inside our bodies to how we might combat climate change.
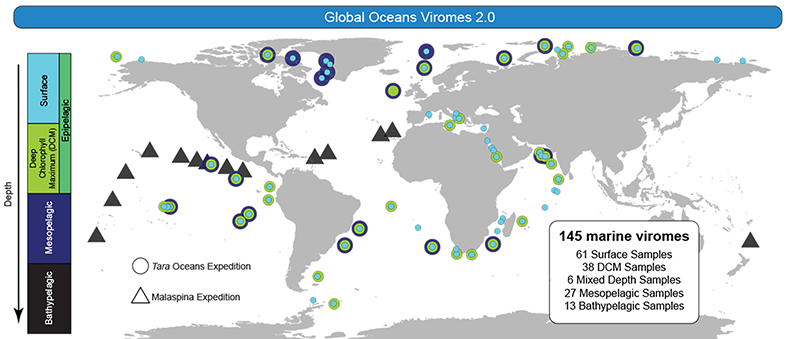
Sullivan’s lab develops datasets and informational tools that help scientists peer into viruses found in soils, oceans, humans and numerous other environments. This year, a computational research scientist in Sullivan’s lab, Benjamin Bolduc, Ph.D., used each of OSC’s three clusters—Pitzer, Owens and Ruby—to create a large-scale analysis of viruses found in thawing Arctic soils and the global oceans. The research began when Sullivan was part of the three-year Tara Oceans Expedition, in which more than 200 experts ventured into the world’s oceans, including tropical, temperate, Antarctic and Arctic waters, to collect samples of the many ocean inhabitants, including viruses and bacteria.
He and Bolduc then harnessed the high performance computing power at OSC to conduct two studies.
“Both studies provide critical baselines for scientists to try to evaluate the role of soils and the oceans in climate change as well as a roadmap for how to study viruses in complex communities,” said Sullivan, a professor of microbiology and civil, environmental and geodetic engineering at The Ohio State University.
The first study examined thawing soils from northern latitudes. While the larger group from the expedition is working to discover how microbes are responding to this thaw, Sullivan assessed how viruses in these soils might be involved.
“We found that there were thousands of viruses,” he said, “that they infected many key carbon cycling microbes, and that they contained genes that were critical to carbon cycling metabolisms.”
The second study focused on viral communities in the global oceans, where the more than 200,000 new viral species were discovered and mapped at latitude and depth.
“This highlighted the Arctic as a biodiversity hotspot and provided the first look at diversity within the species as opposed to just studying the number of species,” Sullivan said. “This foundational look at viral ecology in the oceans will provide baselines for countless future studies.”
Sullivan’s lab used microbiome apps that his team either developed or brought to OSC. These apps take terabytes of raw sequence data and help Sullivan develop new methods to simplify the data. The result was what he calls an ecogenomic toolkit—for each virus (“iVirus”) and microbe (“iMicrobe”)—and both toolkits are available at OSC.
“It’s a best-in-the-world type toolkit, and we could not even put these datasets together without the power of OSC,” Sullivan said.
___________
PROJECT LEAD // Matthew Sullivan, Ph.D., The Ohio State University
RESEARCH TITLE // Ecological impacts and drivers of viral communities in the global oceans
FUNDING SOURCES // The Ohio State University, National Science Foundation
WEBSITE // u.osu.edu/viruslab