Search our client documentation below, optionally filtered by one or more systems.
Search Documentation
Search Documentation
Bowtie is an ultrafast, memory-efficient short read aligner. It aligns short DNA sequences (reads) to the human genome at a rate of over 25 million 35-bp reads per hour. Bowtie indexes the genome with a Burrows-Wheeler index to keep its memory footprint small: typically about 2.2 GB for the human genome (2.9 GB for paired-end).
Collectively, the bedtools utilities are a swiss-army knife of tools for a wide-range of genomics analysis tasks. The most widely-used tools enable genome arithmetic: that is, set theory on the genome. While each individual tool is designed to do a relatively simple task, quite sophisticated analyses can be conducted by combining multiple bedtools operations on the UNIX command line.
While we provide a number of Perl modules, you may need a module we do not provide. If it is a commonly used module, or one that is particularly difficult to compile, you can contact OSC Help for assistance, but we have provided an example below showing how to build and install your own Perl modules. Note, these instructions use "bash" shell syntax; this is our default shell, but if you are using something else (csh, tcsh, etc), some of the syntax may be different.
NWChem aims to provide its users with computational chemistry tools that are scalable both in their ability to treat large scientific computational chemistry problems efficiently, and in their use of available parallel computing resources from high-performance parallel supercomputers to conventional workstation clusters.
For more information about citations of OSC, visit https://www.osc.edu/citation.
To cite Owens, please use the following Archival Resource Key:
ark:/19495/hpc6h5b1
Please adjust this citation to fit the citation style guidelines required.
An eligible principal investigator (PI) heads a project account and can authorize/remove user accounts under the project account (please check our Allocations and Accounts documentation for more details). This document shows you how to identify users on a project account and check the status of each user.
Glenn Cluster Retired: March 24, 2016
The Glenn Cluster supercomputer has been retired from service to make way for a much more powerful new system. As an OSC client, the removal of the cluster will result in work being added to the heavy workload being handled by Oakley or Ruby, potentially impacting you.
sbatch messages
shell warning
Submitting a job without specifying the proper shell will return a warning like below:
sbatch: WARNING: Job script lacks first line beginning with #! shell. Injecting '#!/bin/bash' as first line of job script.
Errors
If an error is encountered, the job is rejected.
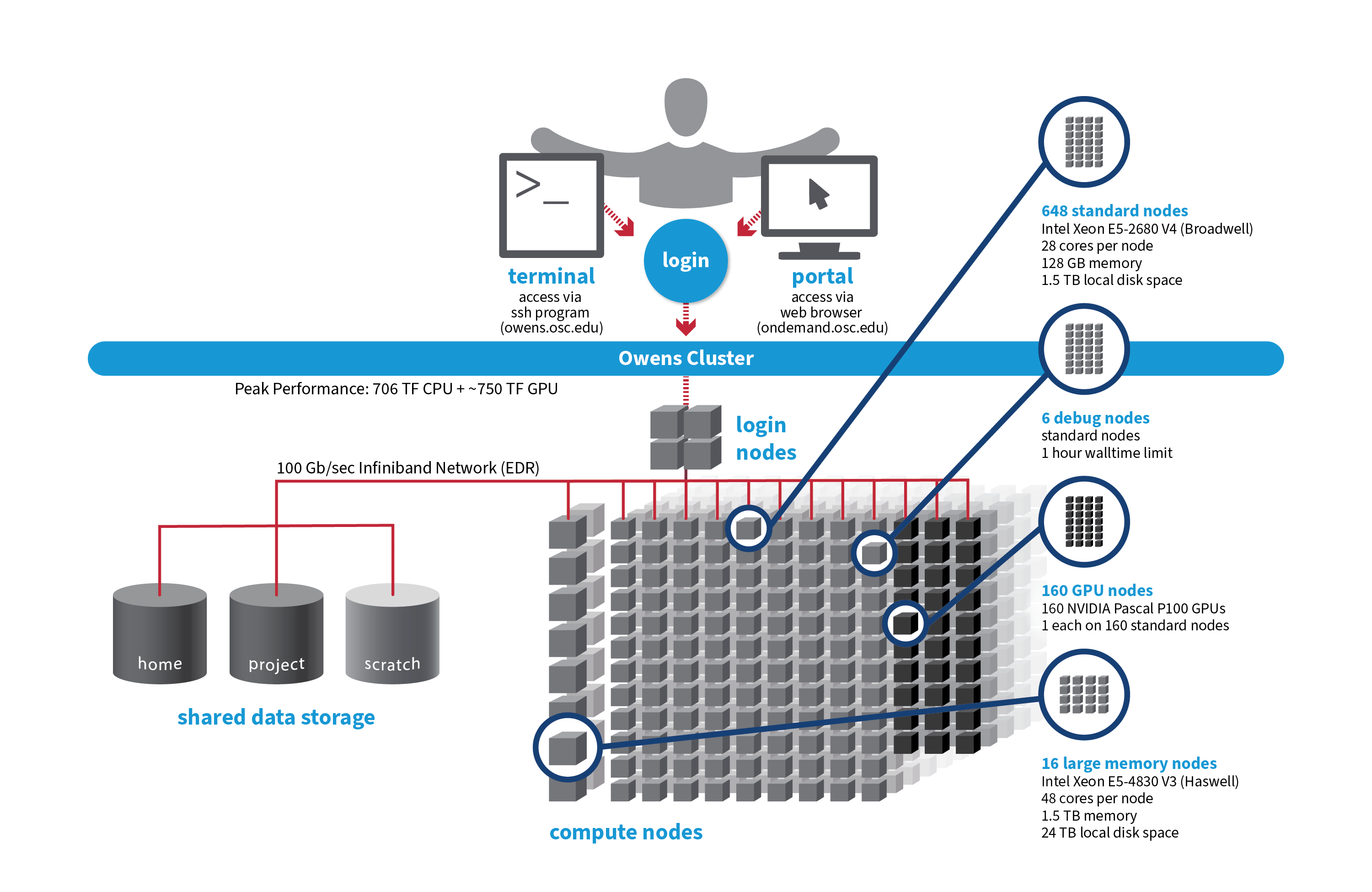
OSC's Owens cluster being installed in 2016 is a Dell-built, Intel® Xeon® processor-based supercomputer.
Linaro MAP is a full scale profiler for HPC programs. We recommend using Linaro MAP after reviewing reports from Linaro Performance Reports. MAP supports pthreads, OpenMP, and MPI software on CPU, GPU, and MIC based architectures.