Search our client documentation below, optionally filtered by one or more systems.
Search Documentation
Search Documentation
sbatch (instead of qsub) command to submit jobs. Refer to the Slurm migration page to understand how to use Slurm. Memory limit
MVAPICH is a standard library for performing parallel processing using a distributed-memory model.
PyTorch Distributed Data Parallel (DDP) is used to speed-up model training time by parallelizing training data across multiple identical model instances.
AutoDock is a a suite of automated docking tools. It is designed to predict how small molecules, such as substrates or drug candidates, bind to a receptor of known 3D structure. AutoDock has applications in X-ray crystallography, structure-based drug design, lead optimization, etc.
rclone is a tool that can be used to upload and download files to a cloud storage (like Microsoft OneDrive) from the command line. It's shipped as a standalone binary, but requires some user configuration before using. In this page, we will provide instructions on how to use rclone to upload data from Google Drive. For instructions with other cloud storage, check rclone Online documentation.
Some projects may wish to have a common account to allow for different privileges than their regular user accounts. These are called community accounts, in that they are shared among multiple users, belong to a project, and may be able to submit jobs. Community accounts are accessed using the sudo command.
A community sudo account has the following characteristics:
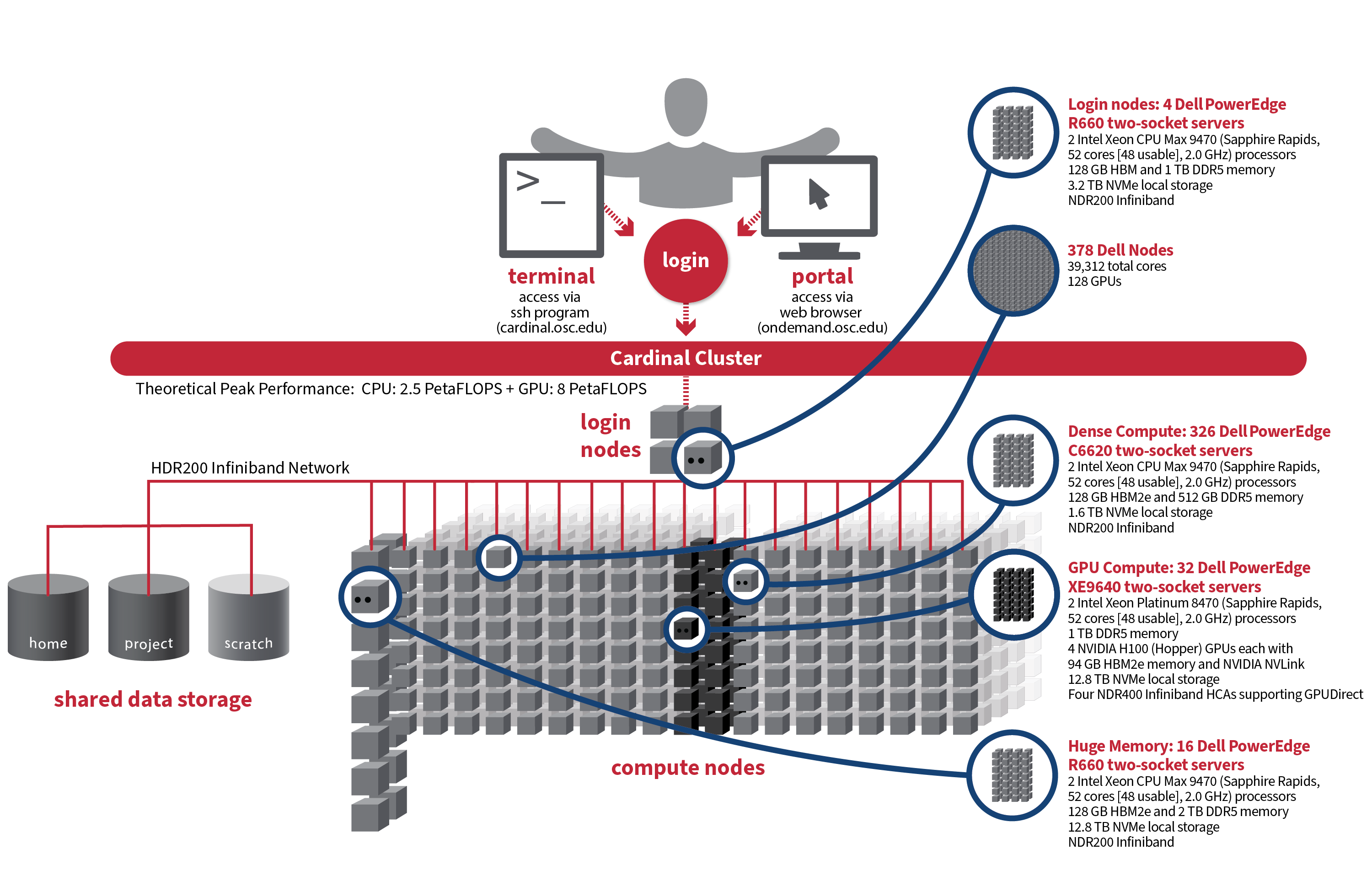
The following are technical specifications for Cardinal.
- Number of Nodes
-
378 nodes
- Number of CPU Sockets
-
756 (2 sockets/node for all nodes)
- Number of CPU Cores
-
39,312
- Cores Per Node
-
104 cores/node for all nodes (96 usable)
Accessing User OneDrive in Globus
Globus is a cloud-based service designed to let users move, share, and discover research data via a single interface, regardless of its location or number of files or size.